Design of 1-(furan-2-yl)-N-(5-substituted phenyl-1, 3, 4-thiadiazol-2-yl) methanimine derivatives as Enoyl-ACP reductase inhibitors: Synthesis, molecular docking studies and anti-tubercular activity
Abstract
A series of 5-phenylsubstiuted 1, 3, 4 thiadiazoles clubbed with furan moiety (Fa-Fe) by means of azomethine linkage have been synthesized. All the newly synthesized compounds were characterized by IR,1HNMR and Mass analyses. All the synthesized molecules have been predicted as anti-tubercular in nature by PASS in silico approach. In vitro anti-tubercular screening was performed by alamar blue assay method on Mycobacterium tuberculosis H37Rv strain. Among the synthesized derivatives Fb and Fe were active at 3.1 µg/mL against Mycobacterium tuberculosis H37Rv strain. The mechanism of action of the active compounds was carried out by docking of receptor enoyl-ACP reductase. It has been concluded that both Fb and Fe posses a significant interaction of hydrogen bonding and electrostatic attraction with Tyr 158 and Met103 in the active site of enzyme.
Introduction
Tuberculosis (TB), a lung infection is one of the contagious and deadly diseases which have added to the woes of the mankind. It is the most dreadful infectious disease caused by Mycobacterium tuberculosis. The widespread of this disease is primarily due to the population growth, emergence of multi-drug resistant TB strains, financial burden in the developing countries and unsuccessful attempt to synthesize a new drug with novel mechanism of action (Thomas et al., 2011). Mycolic acid biosynthesis is essential for the building up of cell wall in mycobacterial and related species (Kolattukudy et al., 1997). InhA, the enoyl acyl carrier protein reductase from Mycobacterium tuberculosis is one of the key enzymes involved in the mycobacterial fatty acid elongation cycle and has been validated as an effective antimicrobial target (Banerjee et al., 1994). A series of pyrrolidine carboxamides have been recognized as potent direct class of InhA inhibitors (He et al., 2006). Recently Oxadiazolo pyrrolidine carboxamides were designed as enol-ACP reductase inhibitors (Sonia and Ravi, 2013). Considering the structural features of the inhibitor of enol-ACP reductase enzyme, we had undertaken the design a novel class of 1,3,4-thiadiazole clubbed with the furan moiety by means an azomethine linkage (Figure 1).
Figure 1: Design of enoyl-ACP reductase inhibitor based up on known pyrrolidine carboxamides
The Schiff base associated with the 2-furfural exhibit antimicrobial and antiproliferative properties were already reported (Gaballa et al., 2007; Rajendran et al., 2010; Hranjec et al., 2011). 1,3,4-thiadiazole pharmacophore displayed a wide range of pharmacological activities such as anti-inflammatory, CNS depressant activity and mucomembranous protector (Varandas et al., 2005; Jatav et al., 2008; Mathew et al., 2013). It has been predicted by the PASS computational approach, that the combination of above these pharmacophore in the designed scaffold would exhibit better anti-tubercular nature.
Materials and Methods
Melting points were determined by using open capillary tube method and the values were uncorrected. IR spectra were recorded on JASCO FT/IR-140 spectrophotometer by using KBr pellets technique. PMR spectra were recorded using BRUCKER FT-NMR-500 MHz spectrophotometer by using DMSO as solvent and TMS as internal standard. The chemical shift was expressed in d ppm. Mass spectra were recorded on a JEOL GCmate mass spectrometer.
Synthesis of thiosemicarbazones
Aromatic aldehyde (0.2 M) in warm alcohol (300 mL) and thiosemicarbazide (0.2 M) in warm water (300 mL) were mixed slowly with continuous stirring. The product separated immediately on cooling which was filtered with suction, dried and recrystallized in ethanol: water (7:3).
Synthesis of 5-substituted phenyl-1, 3, 4-thiadiazol-2-amines
Thiosemicarbazone (0.05 M) was suspended in 300 mL warm water, FeCl3 (0.15 M) in 300 mL water was added quantitatively, slowly with constant stirring. The contents were heated at 80-90°C for 45 min. Solution was filtered hot and then citric acid (0.11 M) and sodium citrate (0.05 M) were added. The resulting mixture was divided into 4 parts and each part was neutralized separately with ammonia (10%). The required amine separated out, filtered with suction, dried and recrystallized in ethanol:water (5:5).
Synthesis of 1-(furan-2-yl)-N-(5-substiuted-phenyl-1, 3, 4-thiadiazol-2-yl) methanimine
5- phenyl-1, 3, 4-thiadiazol-2-amine (0.01M) was suspended in DMF and add furan-aldehyde (0.015 M) with 2-3 drops of Conc. H2SO4.The reaction mixture was then refluxed for 6-7 hours. The reaction progress was monitored by TLC (chloroform:ethanol 4:1). The resultant contents were poured into crushed ice. The crude product was filtered, washed with water until it is free from acidic catalyst, dried and recrystallized with ethanol.
1-(furan-2-yl)-N-(5-phenyl-1, 3, 4-thiadiazol-2-yl) methanimine (Fa): M.p-175-178, Rf: 0.65 IR: vmax /cm-13160(-CH aryl), 1615(C=N), 732(C-S-C). 1HNMR (DMSO-d6/TMS): 8.46(s, 1H, N=CH), 7.21-7.96(m, 8H, aryl protons).MS: m/z (M+)+255.
N-[5-(4-chlorophenyl)-1, 3, 4-thiadiazol-2-yl]-1-(furan-2-yl) methanimine (Fb): M.p-170-172, Rf: 0.72, IR: vmax /cm-13010(-CH aryl), 1588(C=N), 727(C-S-C). 1HNMR (DMSO-d6/TMS):8.23(s, 1H, N=CH), 7.19-7.78(m, 7H, aryl protons). MS: m/z (M+2)+289.
1-(furan-2-yl)-N-[5-(4-methoxyphenyl)-1, 3, 4-thiadiazol-2-yl] methanimine (Fc): M.p-183-186, Rf: 0.81, IR: vmax /cm-13150(-CH aryl), 1595(C=N), 742(C-S-C). ). 1HNMR (DMSO-d6/TMS):8.55(s, 1H, N=CH), 7.01-7.90(m, 7H, aryl protons), 3.55(s, 3H, OCH3). MS: m/z (M+)+285.
1-(furan-2-yl)-N-[5-(2-nitrophenyl)-1, 3, 4-thiadiazol-2-yl] methanimine (Fd): M.p-165-168 Rf: 0.75, IR: vmax /cm-13133(-CH aryl), 1625(C=N), 728(C-S-C). IR: vmax /cm-13176(-CH aryl), 1610(C=N), 735(C-S-C). 1HNMR (DMSO-d6/TMS):8.87(s, 1H, N=CH), 7.2-7.95(m, 7H, aryl protons) MS: m/z (M+1)+300.
1-(furan-2-yl)-N-[5-(3-nitrophenyl)-1, 3, 4-thiadiazol-2-yl] methanimine (Fe): M.p-165-168 Rf: 0.75, IR: vmax /cm-13165(-CH aryl), 1623(C=N), 721(C-S-C). 1HNMR (DMSO-d6/TMS):8.74(s, 1H, N=CH), 7.1-8.01(m, 7H, aryl protons) MS: m/z (M+1)+300.
Computational studies
PASS prediction
Prediction of this spectrum by PASS is based on SAR analysis of the training set containing more than 35,000 compounds which have more than 500 kinds of biological activity. If Pa>0.7 the chance to find the activity in experiment is high, but in many cases the compound may occur to be the close analogue of known pharmaceutical agents. If 0.5<Pa<0.7 the chance to find the activity in experiment is less, but the compound is not so similar to known pharmaceutical agents. If Pa<0.5 the chance to find the activity in experiment is even more less, but if it will be confirmed the compound might occur to be a New Chemical Entity. It was shown that the numbers of experiments necessary to find the known effects for the heterogenous set of substances by using PASS prediction and without it are 1650 and 316 respectively. (Poroikov and Filimonov, 2002; Mathew et al., 2016).
Molecular docking methodology
Preparation of proteins and ligands
The crystal structure of Enoyl Acyl Carrier Protein Reductase (InhA) from Mycobacterium tuberculosis (PDB entry: 2H7M) was retrieved from the Protein Data Bank (http://www.rcsb.org). The protein is refined by removing the inhibitor and water molecules. Hydrogen atoms were added for correct the tautomeric and ionization states of amino acid residues. The energy minimization of the refined protein was done by energy minimizing tool of UFF field. Then the modified protein structure obtained was saved in pdb format and used for all further docking studies. All the final synthesized derivatives, inhibitors and standards were built by using ACDLABS ChemSketch 12.0 version software. The obtained structures were saved in mol format and can be imported in to the workspace of Argus lab 4.0.1.version. The geometry optimization was done by using UFF molecular mechanics method. Clean hybridization option was done and make sure with exact hybridization pattern of the molecule. Final geometry optimization was done by using semi empirical Quantum mechanics PM3 method. The energy minimized structures were saved in pdb format for further docking studies. (Mathew et al., 2016).
Determination of the active site of the enzyme
The active sites of the (PDB entry: 2HYM) were identified by using Q-Site Finder: an energy-based method for the prediction of protein-ligand binding sites (Laurie et al., 2005). The predicted sites comprised of ILE 21, MET 103, GLY 104, MET 147, ASP 148, PHE 149, MET 155, PRO 156, ALA 157, TYR 158, LYS 165, VAL 189, ALA 191, GLY 192, PRO 193, ILE 194, THR 196, MET 199, ILE 202, LEU 207 were the interacting residues. These predicted amino acid residues were selected and saved as the binding site for the docking study of final designed derivatives.
Docking calculation
Argus lab 4.0.1.version was employed in the docking between protein and ligand using argus dock with a fast and simplified potential of mean force (PMF). Dock a ligand in to binding site was done by "Argus Dock" as the docking engine. "Regular precision" was selected in docking precision menu, "Dock" was selected as calculation type, "Flexible" for the ligand and "Ascore" was used for the scoring function. The binding site box was set in to (24 x 22 x 22) angstroms for enclosing the entire active binding site of the protein with a grid resolution of 0.4Ã…. The process of docking is repeated until a constant value of docking score is obtained. The resulted final docked structures were saved in pdb format and the docking snap shots were imported in to Molegro virtual viewer version 2012 2.5.0. This program can generate the amino acid residue around each segment of the final synthesized derivatives and interpreting the different drug-receptor interaction such as hydrogen bonding, hydrophobic interaction and electrostatic force of attraction.
Validation of potential means force method
It is necessary to perform the docking of 1-cyclohexyl-N-(3,5-dichlorophenyl)-5-oxopyrrolidine-3-carboxamide into the active site of enoyl-ACP reductase (2H7M), before docking the test compounds for validate the molecular docking program prescribed in the current study. This inhibitor of binds in the active site region of enoyl-ACP reductase with a binding score of -11.7 with a root mean square of 0.23 was observed.105 number of final unique configurations appeared during the docking calculation. The major interaction of the inhibitor is shown in (Figure 2).
Figure 2: 2D Binding interaction of the inhibitor in the active site enoyl-ACP reductase
Anti-tubercular activity
The antimycobacterial activity of synthesized compounds (Fa-Fe) were assessed against M. tuberculosis using microplate Alamar Blue assay (MABA). This methodology is non-toxic, uses a thermally stable reagent and shows good correlation with proportional and BACTEC radiometric method. Briefly, 200 µL l of sterile deionzed water was added to all outer perimeter wells of sterile 96 wells plate to minimized evaporation of medium in the test wells during incubation. The 96 wells plate received 100 µL of the Middlebrook 7H9 broth and serial dilution of compounds were made directly on plate. The final drug concentrations tested were 100 to 0.2 µg/mL. Plates were covered and sealed with parafilm and incubated at 37ºC for five days. After this time, 25 µL of freshly prepared 1:1 mixture of Almar Blue reagent and 10% tween 80 was added to the plate and incubated for 24 hours. A blue colour in the well was interpreted as no bacterial growth, and pink colour was scored as growth. The MIC was defined as lowest drug concentration which prevented the colour change from blue to pink (Collins and Franzblau, 1997).
Result and Discussion
The synthetic strategy involves in the formation 1-(furan-2-yl)-N-(5-substituted phenyl-1, 3, 4-thiadiazol-2-yl) methanimines (Fa-Fe) was accomplished in three steps and the steps were outlined in (Figure 3).
Figure 3: Synthetic route of the titled compounds
Thiosemicarbazone was obtained by the reaction between aromatic aldehyde and thiosemicarbazide. This thiosemicarbazones undergo oxidative cyclization with FeCl3 in presence of citric acid medium to form 5-phenyl 2-amino 1,3,4-thiadiazole. The amino group of the thiadiazole ring undergoes nucleophilic addition followed by dehydration with furfural in presence of conc.H2SO4 with DMF medium (Mathew et al., 2011). The presence of a band at 1618 cm-1 in Fc strongly recommend the presence of azomethine linkage. A band of 721 and 1350 cm-1 indicated the presence of C-S-C and C-O groups. In 1HNMR a singlet peak at 8.55 showed the presence of N=CH unit in Fc. The deshielding of this CH signal is due to the strong inductive effect of neighbouring electronegative nitrogen atom. The appearance of a singlet peak in the region of 3.55 showed the presence of CH3 group in the methoxyl group of Fc. The multiplet peaks ranges from 7.0-7.90 revealed the presence of aryl protons in the final synthesised derivative. The mass fragment of Fe showed a molecular ion peak M+ at m/z 300 correspond to molecular formula C13H8N4O3S.A peak of m/z 301 of M++1 was also observed. The splitting of a oxygen atom group from the Fe derivative showed a peak of 285 which correspond to the molecular formula of C13H8N4O2S gave a full agreement of the incorporation of the imines linkage between furfural and 2-amino 5(3-nitro phenyl) 1,3,4-thiadiazole. 254, 233, 206, 106 and 94 are the other prominent peaks obtained. The mass fragmentation pattern of Fe is shown in (Figure 4).
Figure 4: Mass fragmentation pattern of Fe
The predicted activity spectra of the 1-(furan-2-yl)-N-(5-substiuted-phenyl-1,3,4-thiadiazol-2-yl) methanimines (Fa-Fe) in the current study was done to identify its anti-tubercular nature with a Pa scores more than 0.68. The activity score of the all designed molecules were shown in Table I.
Table I: PASS prediction of Fa-Fe
| Compound code | Anti-tubercular activity | Anti-tubercular activity |
|---|---|---|
| Pa | Pi | |
| Fa | 0.710 | 0.004 |
| Fb | 0.691 | 0.004 |
| Fc | 0.689 | 0.004 |
| Fd | 0.709 | 0.004 |
| Fe | 0.804 | 0.003 |
The prediction of the pharmacological profile was established by the in vitro studies. The rank order of PASS showed as Fe>Fa>Fd >Fb>Fc. The PASS program significantly predicted the Fe compound has the highest probability and was most active in the in vitro anti-tubercular studies, while Fc with lowest Pa value in the series was practically less active. It has been noted that the PASS program predicts Fb with a low Pa value of 0.691. This low score cannot favours its activity ratio because it showed significant activity with MIC of 3.3 µg/mL.
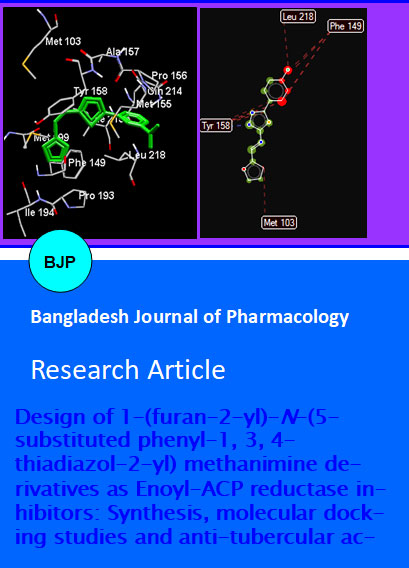
The active pocket was considered to be the site where 1-cyclohexyl-N-(3, 5- dichlorophenyl)-5-oxopyrrolidine-3-carboxamide (pyrrolidine carboxamides) complexed with enoyl-ACP reductase in 2H7M. Hydrogen bond-ing network of ligand and Tyr158 seems to be a prominent feature among all the InhA-inhibitor complexes identified so far (Kuo et al., 2003). All the newly designed molecules showed good binding interaction towards the active site of enoyl-ACP reductase. Molecular docking study suggested that compound Fb and Fe showed the best docking score of-9.2778 and 9.2311 respectively. The docking pose of Fe showed a crucial π-π stacking interaction between the azomethine unit and phenyl system of Tyr158 (Figure 5).
Figure 5: Docking of Fe into the active site of enoyl-ACP reductase
The in silico study of the Fb and Fe revealed that its binding affinity towards the enzyme is due to the hydrogen bonding interaction with Tyr 158.The ligand map of Fb (Figure 6) suggested that a steric interaction of furan part in the scaffold to the Met 103 also contribute an interesting result in the activity ratio. Tyr 158 and Met 103 are the major interacting residues of the known inhibitor of 1-cyclohexyl-N-(3,5-dichlorophenyl)-5-oxopyrrolidine-3-carboxamide towards enoyl-ACP reductase.
Figure 6: 2D-ligand plot of Fb with significant interaction with Tyr 158 and Met103
Another interesting factor raised from our finding is that the poor affinity and MIC of Fc is due to its lack of hydrogen bonding between Tyr158. Estimated binding energy (Kcal/mol) of all the designed molecules and the amino acid residues enveloped by the designed molecules was represented in Table II.
Table II: Molecular docking score of (Fa-Ff) in the active site of enoyl-ACP reductase
| Compound code | Binding score (Kcal/mol) | No. of Hydrogen bonds | Amino acid residues enveloped |
|---|---|---|---|
| Fa | -8.7903 | 1 | Phe149, Tyr158, Met155, Pro156, Met199, Gln 214 |
| Fb | -9.2778 | 2 | Met103, Phe149, Tyr158, Leu218 |
| Fc | -8.4321 | 2 | Tyr-158, Pro156, Met199, Gln214 |
| Fd | -8.8783 | 0 | Tyr 158, Phe 149, Met155, Gln214 |
| Fe | -9.2311 | 1 | Phe149, Met 155, Tyr 158, Pro156, Gln214, Leu218 |
Anti-tubercular screening
Compounds Fe and Fb showed significant activity with MIC of 3.1 µg/mL. It is interesting to note that the activity ratio is decreased in the designed structures when the presence of electron donating group such as methoxy group in the phenyl system. The promising activity of the designed molecules is mainly attributed with the presence of electron withdrawing group such as nitro and chloro in the phenyl system.2-nitro (Fd) also showed a comparable MIC of 12.5 µg/mL.
The present study established the design and synthesis of some new phenyl substituted 1,3,4-thiadiazole clubbed with furan moiety as a potent inhibitor of enoyl-ACP reductase. The anti-tubercular activity of the designed scaffold was predicted by PASS in silico approach. Molecular docking study revealed that the structural features of the derivatives can make a significant interaction towards the active site of the enzyme which is necessary for the development of mycolic acid. In the light of our interesting results of some of the derivative's binding mode, a new design has been developed for the inhibition of enoyl-ACP reductase.
Acknowledgements
The authors are highly thankful to Maratha Mandal's Nathajirao G. Halgekar Institute of Dental Sciences and Research Centre; Belgaum for anti-tubercular activity. Our sincere thanks goes to Grace College of Pharmacy, Kerala, India, for carrying out the synthetic work for the present study. Authors also acknowledge the, SAIF IIT, Chennai, for carrying out the spectral analysis.
References
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR Jr. InhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 1994; 263: 227-30.
Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997; 41: 1004-09.
Gaballa AS, Asker MS, Barakat AS, Teleb SM. Synthesis, characterization and biological activity of some platinum (II) complexes with Schiff bases derived from salicylaldehyde, 2-furaldehyde and phenylenediamine. Spectrochim Acta A. 2007; 67: 114-21.
Hranjec M, StarÄević K, Pavelić SK, LuÄin P, Pavelić K, Zamola GK. Synthesis, spectroscopic characterization and anti proliferative evaluation in vitro of novel Schiff bases related to benzimidazoles. Eur J Med Chem. 2011; 46: 2274-79.
He X, Alian A, Stroud R, Ortiz de Montellano PR. Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis. J Med Chem. 2006; 49: 6308-23.
Jatav V, Mishra P, Kashaw S, Stables JP. Synthesis and CNS depressant activity of some novel 3-[5-substituted1, 3, 4-thiadiazole-2-yl]-2-styryl quinazoline-4(3H)-ones. Eur J Med Chem. 2008; 43: 135-41.
Kolattukudy PE, Fernandes ND, Azad AK, Fitzmaurice AM, Sirakova TD. Biochemistry and molecular genetics of cell-wall lipid biosynthesis in mycobacteria. Mol Microbiol. 1997; 24: 263-70.
Kuo MR, Morbidoni HR, Alland D, Sneddon SF, Gourlie BB, Staveski MM, Leonard M, Gregory JS, Janjigian AD, Yee C, Musser JM, Kreiswirth B, Iwamoto H, Perozzo R, Jacobs WRJr, Sacchettini JC, Fidock DA. Targeting tuberculosis and malaria through inhibition of enoyl reductase: Compound activity and structural data. J Biol Chem. 2003; 278: 20851-59.
Laurie AT, Jackson RM. Bioinformatics. Q-SiteFinder: An energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 2005; 21: 1908-16.
Mathew B, Suresh AJ, Mathew GE, Shankar S, Kumar SS. Synthesis, characterisation, and anti microbial screening of some 2-amino,5-(phenyl substituted)1,3,4-thiadiazole derivatives. Int J Chem Tech Res. 2011; 3: 364-68.
Mathew B, Suresh J, Anbazhagan S. Development of novel (1-H) benzimidazole bearing pyrimidinetrione based MAO-A inhibitors: Synthesis, docking studies and antidepressant activity. J Saudi Chem Soc. 2016; 20: S132-39.
Mathew B, Suresh J, Anbazhagan S. Synthesis and PASS-assisted in silico approach of some novel 2-substituted benzimidazole bearing a pyrimidine-2, 4, 6(trione) system as mucomembranous protector. J Pharm Bioallied Sci. 2013; 5: 39-43.
Mathew B, Suresh J, Anbazhagan S, Chidambaranathan N. Discovery of some novel imines of 2-amino, 5-thio, 1, 3, 4-thiadiazole as mucomembranous protector. Synthesis, antioxidant activity and in silico PASS approach. J Saudi Chem Soc. 2016: 20: S426-32.
Poroikov VV, Filimonov DA. How to acquire new biological activities in old compounds by computer prediction? J Comput Aided Mol Des. 2002; 16: 819-24.
Rajendran G, Amritha CS, Anto RJ, Cheriyan VT. Synthesis, thermal and anti-tumour studies of Th(IV) complexes with furan-2-carboxaldehyde4-phenyl-3-thiosemicarbazone. J Serb Chem Soc. 2010; 75: 749-61.
Sonia G, Ravi TK. Oxadiazolo pyrrolidine carboxamides as enoyl-ACP reductase inhibitors: Design, synthesis and anti-tubercular activity screening. Med Chem Res. 2013; 22: 3428-33.
Thomas KD, Adhikari AV, Chowdhury IH, Sandeep T, Mahmood R, Bhattacharya B, Sumesh E. Design, synthesis and docking studies of quinoline-oxazolidinone hybrid molecules and their anti-tubercular properties. Eur J Med Chem. 2011; 46: 4834-45.
Varandas LS, Fraga CAM, Miranda ALP, Barreiro EJ. Design, synthesis and pharmacological evaluation of new nonsteroidal anti-inflammatory 1,3,4-thiadiazole derivatives. Lett Drug Des Discov. 2005; 2: 62-67.